AlphaFold provides high quality predictions for 3D structures of proteins based solely on amino acid sequences. The algorithm, based on deep learning methods, has excellent agreement with experimental data. The software is public and can be used by any user to predict structures [1].
Proteins are large molecules essential to all organisms. The sequence of amino acids that form a protein determines its three-dimensional structure. Each protein has a unique shape that determines its function. Being able to predict the spatial structure of a protein from its amino acid sequence has been a long-standing challenge. This task is even more challenging for proteins with a non-trivial topologies.
The AlphaLasso is a server detecting any type of lasso entanglement and a database collecting information about complex lasso proteins [2] predicted by AlphaFold. The lasso entanglement arises, when one terminus of a protein backbone pierces through an auxiliary surface of minimal area, spanned on a covalent loop (see Fig. 1).
The server is designed to study protein chains, however one can upload any kind of (bio)polymer. It is equipped with various tools facilitating perceiving the lasso entanglement. In particular, the user can visualize the protein with the minimal surface spanned on a covalent loop.
The database classifies protein chains according to its entanglement type i.e. number and the direction of threading. Based on these results, the lasso type is prescribed to every closed loop and the overal topology (lasso fingerprint) to the protein chain. For further analysis, AlphaLasso also presents many biological and geometrical statistics. The database contains all the protein structures predicted by AlphaFold with high confidence (pLDDT > 70) that have at least one cysteine bridge.
The results are currently undergoing a manual review process to ensure accuracy and relevance. This review aims to filter out any artifacts, providing users with a higher quality of information. By default, users can view both reviewed and unreviewed information, providing a comprehensive array of results. This choice allows for quicker access to information, albeit with the understanding that it may not have been thoroughly reviewed.
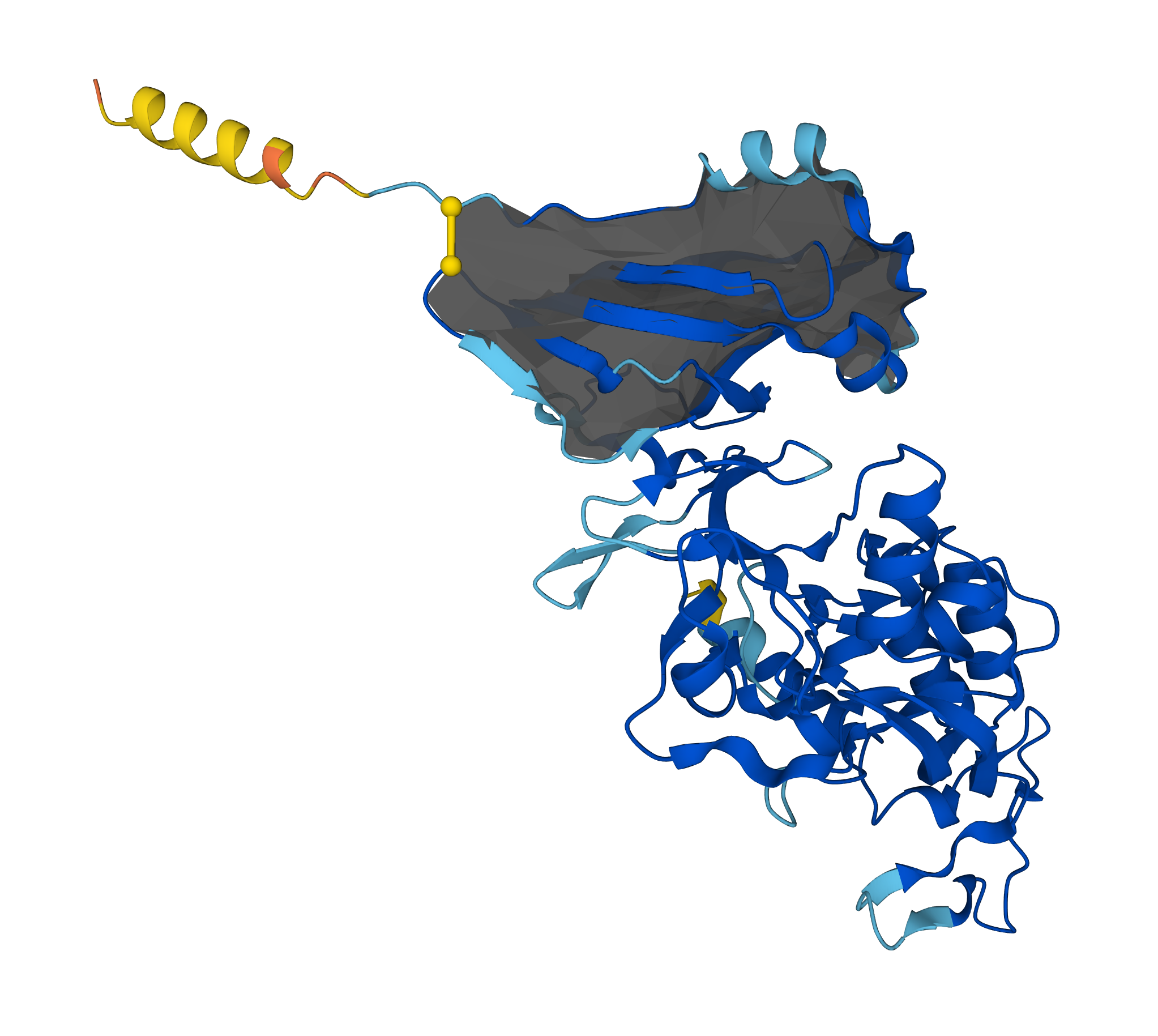
Figure 1. Example of a protein with a lasso L1 (A0A1Z5L3X5).
The first example of complex lasso protein was leptin, in which the loop is closed via cysteine bridge [4-6]. This premier discovery was followed by the thorough study of the non-redundant set of protein chains containing the cysteine-bridge-based loop [2]. This analysis revealed the existence of five distinct major classes, however other classes are possible. The most popular class are the single lasso proteins (L1), in which a covalent loop is pierced by a tail only once. The next are double (L2) and triple (L3) lassos, in which the covalent loop is pierced twice or three times respectively, while after piercing the tail winds back and performes next piercing from the opposite direction. The two remaining classes are two sided-lassos (LLi,j) in which both tails cross the surface i and j times respectively and supercoiling lassos (LS) in which one tail winds around the covalent loop after crossing and then performes piercing from the same direction. These major classes are schematically depicted in Fig. 2. We stress that complex lasso proteins with cysteine loops should not be confused with well known cysteine knot proteins and knottins [7] nor knotted proteins [8]. The complex lasso motif is more general notion than cysteine knots, as cysteine knots require the existence of at least three cysteine loops.
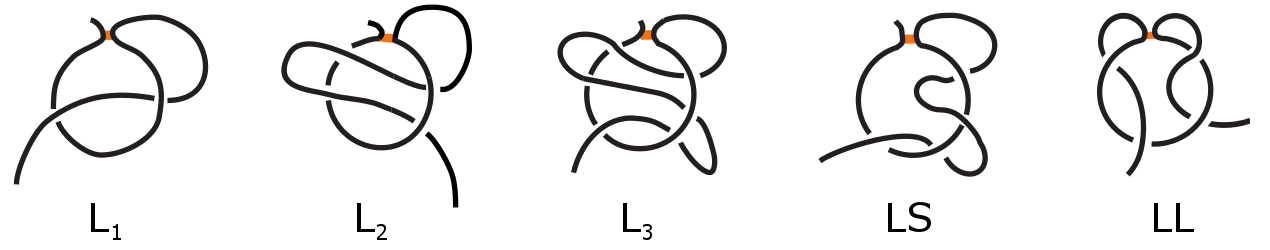
Figure 2. Major classes of complex lasso proteins: single lasso L1, double lasso L2, triple lasso L3, supercoiling LS2 and two-sided lasso LL1,1.
Usually, determination of the threading is impossible by a naked eye. Therefore, we developed [2] the mathematically well defined method which specifies the piercings with its direction unequivocally (see more in the Lasso detection section). This on the other hand enables us to split every major class into smaller subclasses, according to the piercing tail (N- or C-terminal) and the direction of piercing.
Herein, we present the AlphaLasso – the first server and database to analyze loops in AlphaFold-solved protein models while taking into account the pLDDT confidence values. AlphaLasso has two main functions: 1) providing researchers with a webserver for analyzing loops in their own AlphaFold predictions and 2) cataloging loops in AlphaFold predictions for which models have been published.
AlphaLasso server provides a comprehensive topological analysis for single or multiple models uploaded in the CIF or PDB format. The results page shows the lasso types, the lasso pLDDT, an image matrix showing the positions of the loops, a 3D manipulatable model of the protein with colored loop locations and surface, information about the position of lassos along the amino acid sequence, list of homologs, and other information about the protein.
This version of the AlphaLasso database provides information about structures with loops of the whole AlphaFold database: nearly 15M proteins (with chain pLDDT >70) from over 200 million protein structures predicted by AlphaFold. The data is split into two major categories of reviewed and unreviewed structures.
This server and database have been created in a joint collaboration between:
Paweł Rubach1,2, Jacek Płonka1,3, Bartosz Greń1, Fernando Bruno da Silva1, Marta Korpacz1, Joanna I. Sułkowska1,4
The research leading to creation of this database has been supported by the National Science Centre 2022/47/B/NZ1/03480 (to J.I.S.).
The AlphaLasso website is licensed under CC BY 4.0