In AlphaLasso database, the piercing information (described in LassoProt database) obtained for each closed loop in a chain enables us to establish lasso type classification. The loop is closed based on the distance of 3 Å between the sulfur atom, which can form a cysteine bond. In general protein lasso has a loop and two tails (N-tail and C-tail). Rarely, when a cysteine bond is created by the first or last residue there is only one tail. Each tail can pierce a loop, even more then once. We distinguish two sides of the loop and we mark them as + and - side, therefore every tail can be described by a sequence of + and - which describe from which side tail is pinning the loop. Most tails which pierce a loop many times do this in an alternating way forming a +-+-+- or -+-+-+ pattern (order is aligned with residue numbering). However, some of them after piercing wind around a loop and pierce a loop again from the same side, therefore in their pattern are two neighboring pluses or minuses. Such lassos we call supercoiled lassos.
Herein, we take into account the following lasso type as described below: piercings by one tail (supercoiled or not) and two-sided (supercoiled from both sides, from one side, or not supercoiled). Then each type can be divided into sub-categories depending on the number of piercings. In the case of protein predicted based on AlphaFold the complexity of the lasso is much greater than that of the known structures deposited in LassoProt.
Type can be specified further with added piercing pattern e.g. LLS3+,4-++-, which means that from N-tail pierces in an alternating way starting from +, and C-tail pierces from - side, then two times from + side (supercoiling) and again from - side.
In the case when a covalent loop is pierced by one tail, we identified the following topological types: L1–L26, L28, and L30. Note that in this case at least one structure with a given topology has been verified by us. This does not mean that all structures of a given class are correct.
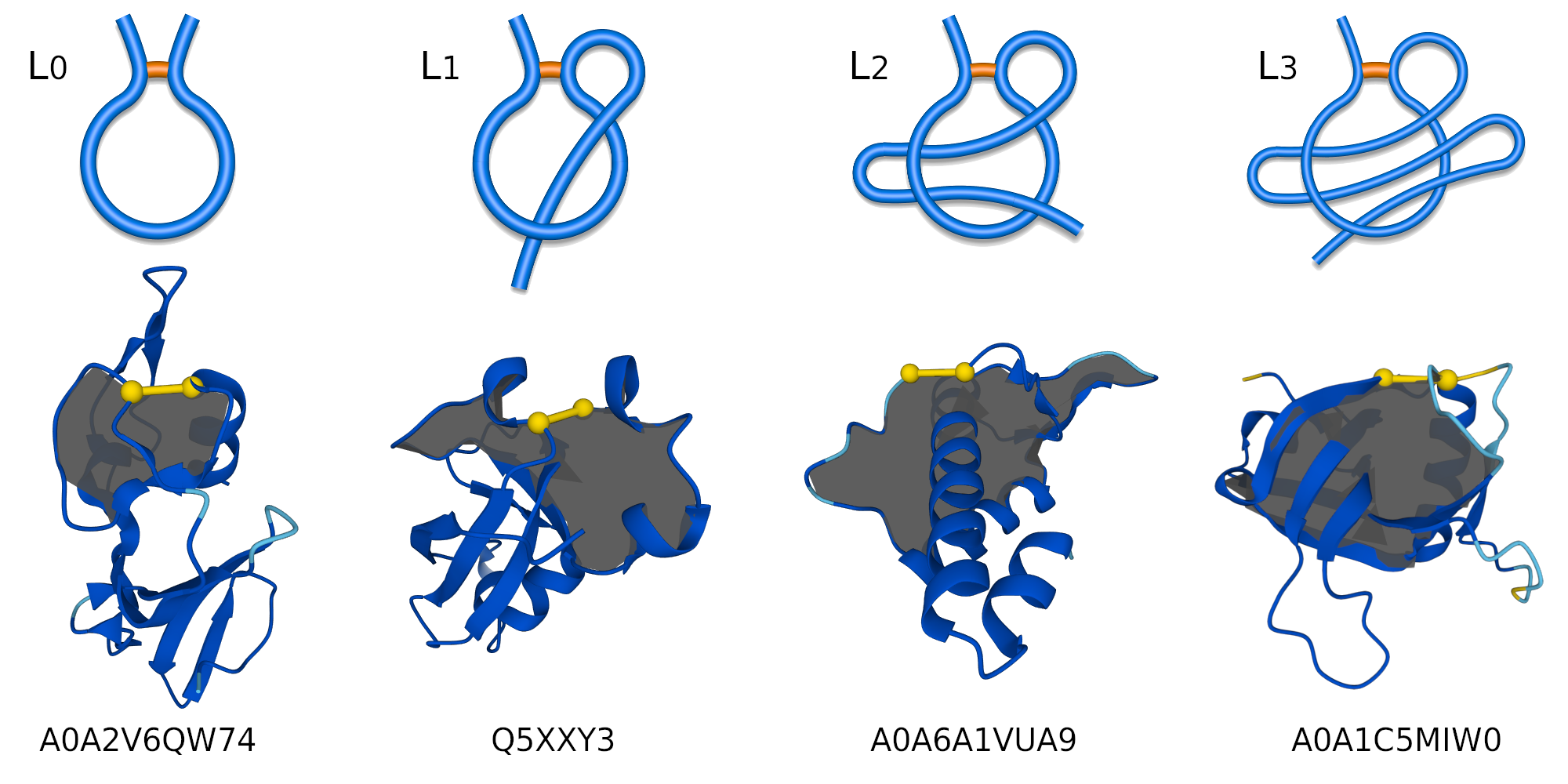
Figure 1. Schematic representation of a regular one-sided lasso, examples of lasso types: L0, L1, L2, and L3 (top panel). Bottom panel, cartoon representations of the protein structures with their AlphaFold IDs.
LL1,1, LL1,2, LL1,3, LL1,4, LL1,5, LL1,6, LL1,7, LL1,8, LL1,9, LL1,11, LL1,12, LL2,1, LL2,2, LL2,3, LL2,4, LL2,5, LL2,6, LL2,7, LL2,8, LL2,9, LL2,11, LL2,15, LL3,1, LL3,2, LL3,3, LL3,4, LL3,5, LL3,6, LL3,7, LL4,1, LL4,2, LL4,3, LL4,4, LL4,5, LL4,6, LL4,9, LL5,1, LL5,2, LL5,3, LL5,4, LL5,5, LL5,6, LL5,7, LL5,9, LL6,1, LL6,2, LL6,3, LL6,4, LL6,5, LL6,8, LL7,1, LL7,2, LL7,5, LL8,2, LL8,5, LL9,1, LL15,1, and LL28,1.
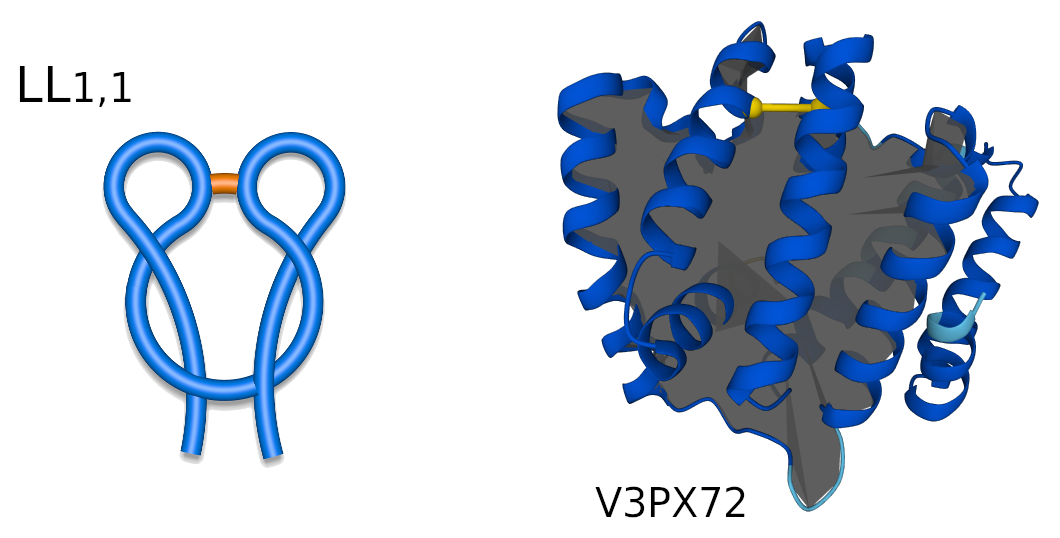
Figure 2. Schematic representation of LL1,1 two-sided lasso (left), and LL1,1 protein structure (right) with its respective AlphaFold ID.
LS2–LS21, LS24, LS26, LS27, LS35, and LS37.
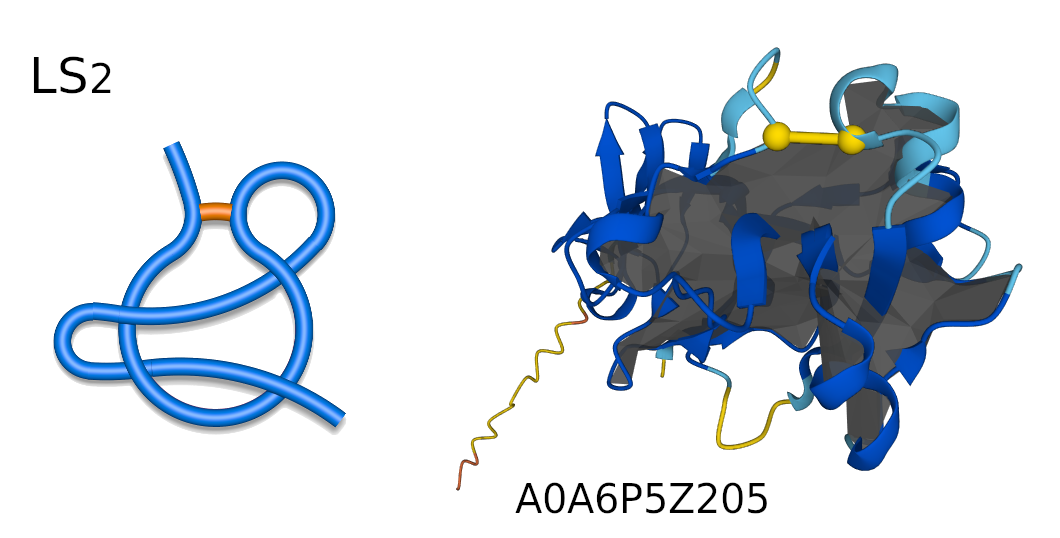
Figure 3. Schematic representation of LS2 supercoiled lasso (left), and LS2 protein structure (right) with its respective AlphaFold ID.
Disulfide bridges, also known as S-S bridges, are strong covalent linkages formed between two cysteine amino acids within a protein, Figure 4. These linkages occur when the side chains of two cysteines, containing thiol groups (SH), react with each other to form a disulfide bond (S-S). Disulfide bridges significantly contribute to protein stability by creating a cross-link between different parts of the protein chain, thereby maintaining its three-dimensional structure.
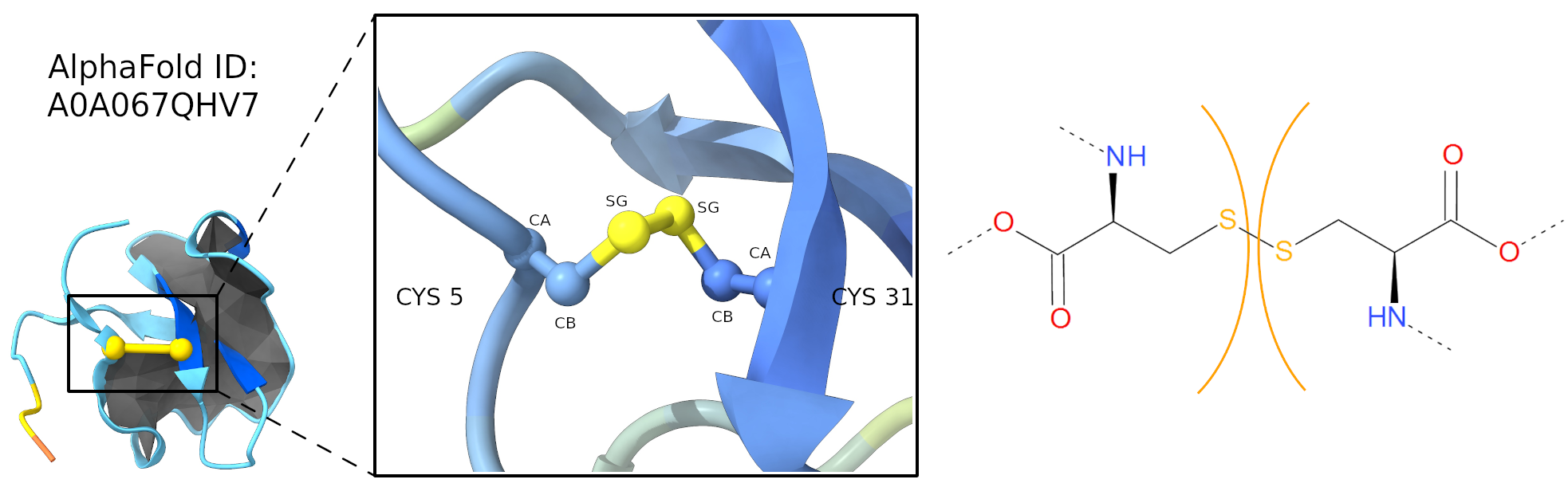
Figure 4. Cysteine (Disulfide) bridge, S-S, in carton (left panel) and stick (right panel) representation. Left panel, cartoon representation of AlphaFold ID A0A067QHV7 protein, lasso type L1, with the minimal surface in dark gray and the cysteine bridge in yellow. The zoom region highlighted the atoms forming the S-S bridge. Right panel, stick representation, the cysteines are separated by orange arcs. Dashed lines denote the remaining part of the protein chain.
The amide category is a short name for all bridges closed by the connection via C-N bond. Apart from most common amide bridges it also contains the amine structures formed by joining two residues. In most cases, the C-N bond is introduced posttranslationally (or aritificially) by the action of specified enzymes. However, autocatalytic mechanism is also possible (as during GFP functional unit formation). An amide bridge arises upon joining free amine and carboxylic groups. The free amine group can be found in side group of lysine and on N-terminus of the protein. The free carboxylic group can be found in the side group of aspartic and glutamic acids as well as on C-terminus of protein. Therefore, the amide linkage occures e.g. when one of the termini is condensed with the interior of the chain, or the termini are joined together (in cyclotide proteins). In the latter case however, one cannot distinguish the termini properly without the "source code" of RNA or DNA which was translated to this protein.
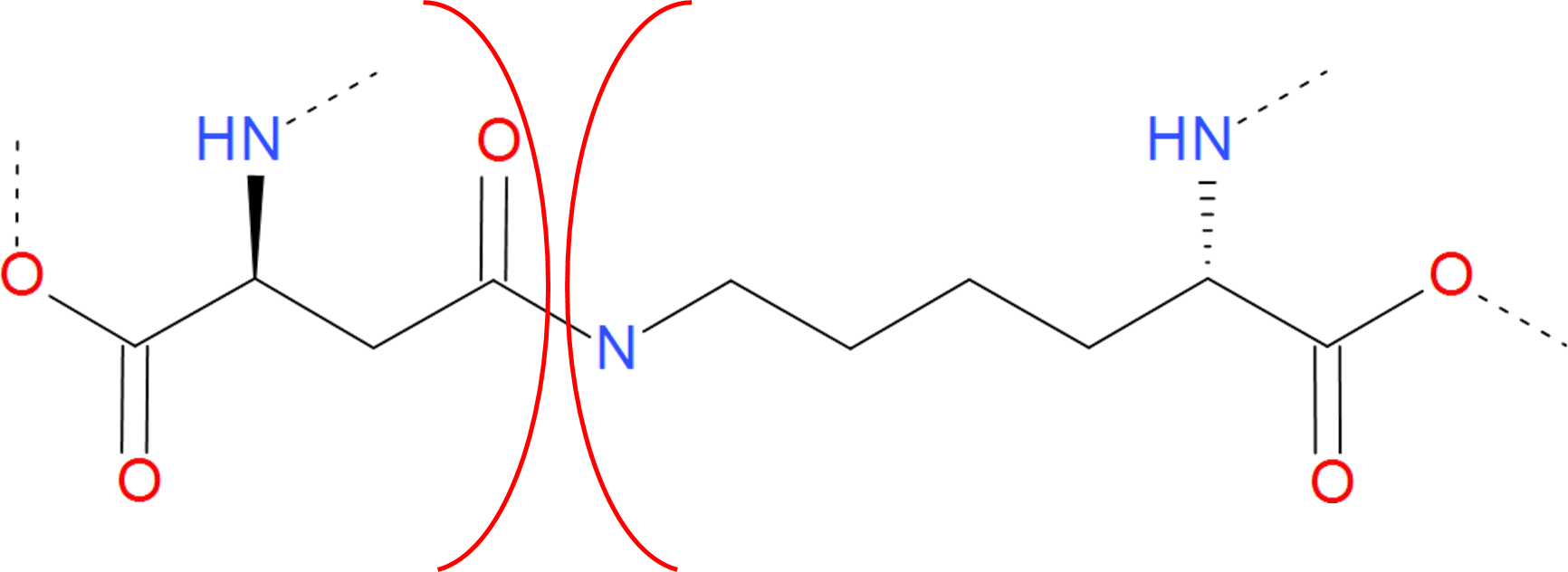
Figure 5. Example of amide bridge between aspartic acid and lysine. The residues are separated by red arcs. The dashed lines denote the rest of the protein chain.
The amide bond can be viewed as the peptide bond, present not in the main chain. Therefore, in proteins it is often called isopeptide bond. Although amide bond is not prone to degradation in reducing/oxidizing conditions, it breaks in low or high pH. The amide bond due to the resonance effect is considered to be partially double. As a result it is stronger and shorter than the e.g. cysteine bond. Moreover, there is no freedom of rotation around the bond axis. The “amine bond” is also formed by joining a carbon and a nitrogen atom. This however, has no keto oxygen (oxygen atom joined by double bond to carbon atom). As a result the electron structure of the bond changes. The bond is single and there is no resonance structure impeding rotation around it. The free electron pair of nitrogen atom is not involved in any resonance structure, therefore the nitrogen atom is still basic (on the contrary, amids are not basic). The bond is not easily broken during hydrolysis. It is therefore more stable, than the amide bond. Similarly, it is formed naturally by the action of specialized enzymes, or autocatalitically. However, as such interactions are in most cases local, the loops introduced by amine bond are often too small and are not taken into our analysis.
The Ester category includes both ester and ether bridges, i.e. bridges characterized by the connection via C-O bond. The ester bridge arises when the free carboxylic group is condensed with hydroxyl group of threonine or serine. As the free carboxylic group is found e.g. on the C-terminus of protein, such bridge can occur upon condensation of C-terminus with the intrachain side group.
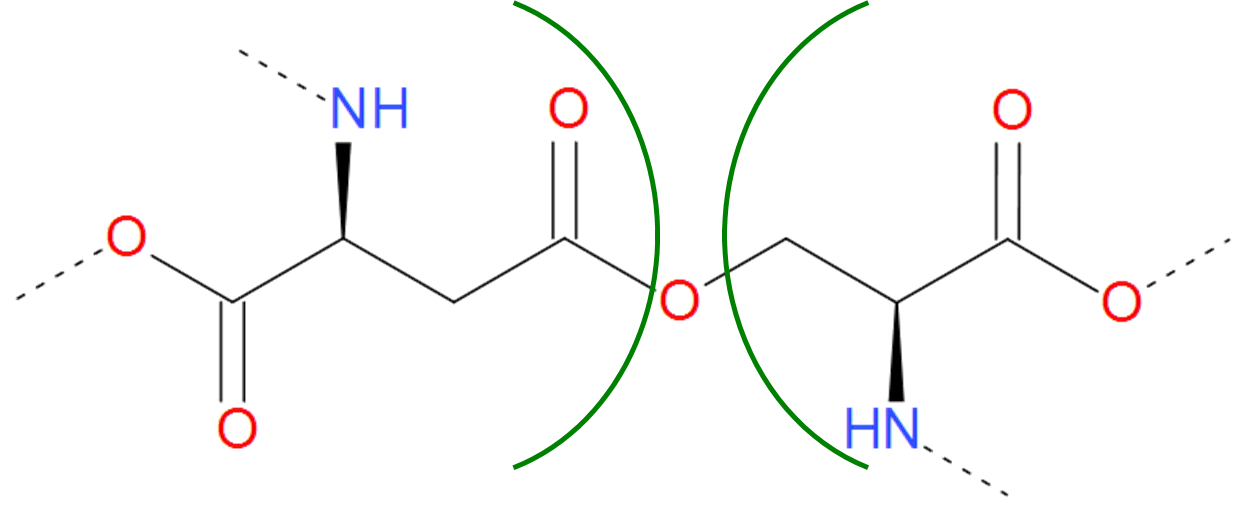
Figure 6.Example of realization of ester bond. The residues of asparaginic acid and serine are separated by green arcs. The dashed lines denote the remaining part of the protein chain.
An ester bond is however more susceptible to hydrolysis than the amide bond. Therefore in some proteins there can be found another realization of C-O bond, namely ether bond. Ether bond is harder to be hydrolyzed, however it is also harder to be synthetized in the cell conditions.
The Thioester category is a sulfuric analog of the Ester category. In contains both thioester and thioether bridges, characterized by the C-S bond, which arrises in much the same way as ester and ether bridges. The only exception is, that instead of serine or threonine, one of the amino acid should be sulfur containing cysteine.
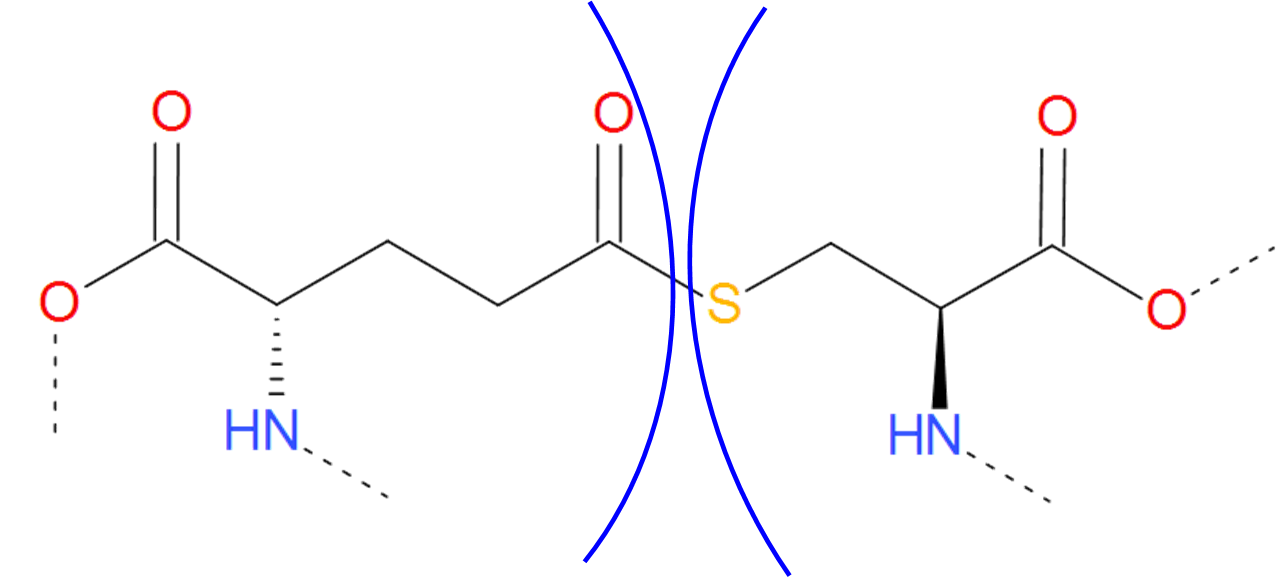
Figure 7.Thioester bridge formed by glutaminic acid and cysteine. The residues are separated by blue arcs. The dashed lines denote the remaining part of the protein chain.
The thioester and thioether are weaker and longer, than their oxygen analogs (ester and ether). Therefore, they can be thought of as a hidden functional cysteine, which can be released in favorable conditions. Such mechanisms could be useful for example in adhesion proteins, which adhere properties would be activated in specified conditions only, and the adhering mechanism would depend on the existence of free thiol (-SH) groups.
Two different file extensions are accepted to compute the lasso properties, CIF and PDB. In a CIF file bridges can be found in the “struct_conn” category. Record describes disulfide bridge if the value of “conn_type_id” is equal to “disulf”. PDB files utilize three different records to identify S-S bridge: (i) "CONNECT” — these lines show which non-backbone atoms are connected within the protein structure; (ii) “SSBOND" or (iii) "LINK" to explicitly define S-S or other bridges.
Unlike structures that can be found in the Protein Data Bank (PDB), AlphaFold generated structures do not contain information on covalent interactions and thus the records mentioned above are empty. However, AlphaLasso doesn't need those records to identify bridges! We conducted an analysis of over 220 thousand structures held in the Protein Data Bank and established a criteria that allows AlphaLasso to automatically find these covalent interactions. We identify:
The AlphaLasso website is licensed under CC BY 4.0